ENSO (cont’d)¶
Here, we will do a similar analysis than last week, but focussing on the equatorial Pacific. The dominant process along the equator is upwelling, due to the westward tradewinds pushing surface waters northward in the northern hemisphere, and southward in the southern hemisphere due to the earth rotation, resulting in upwelling of cold water from below along the equator. While that upwelling is easily seen in satellite sea surface temperature images, can we see it in drifter tracks?
The global database of drifter tracks is widely available over the internet; the instructions on how to obtain the data for any region are given at the end of this lecture. We have extracted the data for the area and saved as a cvs file.
[1]:
import pandas as pd
import numpy as np
drifters = pd.read_csv('/home/ocn463-data/woce/drift/pacdrift.gz', compression='gzip', na_values = -999.99)
[2]:
drifters
[2]:
| id | month | day | year | lat | lon | sst | u | v | umag | latvar | lonvar | tempvar | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 7702986 | 3 | 29.75 | 1988 | 2.373 | 267.958 | 28.244 | -76.816 | 37.069 | 85.293 | 0.000093 | 0.000151 | 0.010206 |
| 1 | 7702986 | 3 | 30.00 | 1988 | 2.442 | 267.809 | 28.281 | -78.719 | 32.167 | 85.037 | 0.000008 | 0.000010 | 0.003204 |
| 2 | 7702986 | 3 | 30.25 | 1988 | 2.499 | 267.652 | 28.063 | -78.133 | 24.650 | 81.929 | 0.000045 | 0.000065 | 0.010097 |
| 3 | 7702986 | 3 | 30.50 | 1988 | 2.538 | 267.505 | 28.070 | -71.856 | 23.063 | 75.467 | 0.000027 | 0.000038 | 0.007007 |
| 4 | 7702986 | 3 | 30.75 | 1988 | 2.589 | 267.373 | 28.302 | -74.005 | 27.462 | 78.936 | 0.000047 | 0.000069 | 0.006161 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2244751 | 90561 | 3 | 30.25 | 2011 | 9.961 | 180.135 | 27.692 | -41.105 | -1.209 | 41.123 | 0.000006 | 0.000008 | 0.004182 |
| 2244752 | 90561 | 3 | 30.50 | 2011 | 9.964 | 180.069 | 27.607 | -37.501 | 3.801 | 37.693 | 0.000003 | 0.000006 | 0.004396 |
| 2244753 | 90561 | 3 | 30.75 | 2011 | 9.975 | 179.988 | 27.594 | -36.597 | 2.026 | 36.653 | 0.000008 | 0.000010 | 0.003982 |
| 2244754 | 90561 | 3 | 31.00 | 2011 | 9.972 | 179.924 | 27.704 | -36.611 | 4.784 | 36.923 | 0.000022 | 0.000027 | 0.004592 |
| 2244755 | 90561 | 3 | 31.25 | 2011 | 9.994 | 179.843 | 27.712 | -36.054 | 11.626 | 37.882 | 0.000008 | 0.000010 | 0.004258 |
2244756 rows × 13 columns
There’s a lot more data here than there was in the previous lecture. Let’s make a plot of every 100th track
[3]:
import matplotlib.pyplot as plt
fig, ax = plt.subplots(figsize=(12,12))
ax.plot(drifters['lon'][::100], drifters['lat'][::100], '.')
ax.set_title('Drifter tracks')
ax.set_aspect('equal')

There are some gaps, but we can still see what the currents are like. Let’s make boxes of 1/2 degree latitude and all longitudes, and find the average U and V in each box as well as the standard deviation of them.
NOTE: The code block below takes about 5 or 6 minutes to run on our server, so let’s NOT ALL RUN AT THE SAME TIME
[4]:
cenlon, cenlat = np.meshgrid(np.arange(165.5, 267.6, 0.5), np.arange(-9.5, 9.6, 0.5))
Umean = np.nan * np.ones_like(cenlon)
Vmean = np.nan * np.ones_like(cenlon)
Ustd = np.nan * np.ones_like(cenlon)
Vstd = np.nan * np.ones_like(cenlon)
for ilat in range(cenlon.shape[0]):
for ilon in range(cenlon.shape[1]):
boxlat = np.where(np.absolute(drifters['lat']-cenlat[ilat, ilon]) < 0.5)[0]
boxlon = np.where(np.absolute(drifters['lon']-cenlon[ilat, ilon]) < 0.5)[0]
box = np.array(sorted(set(boxlat) & set(boxlon)))
Umean[ilat, ilon] = np.nanmean(drifters['u'][box])
Vmean[ilat, ilon] = np.nanmean(drifters['v'][box])
Ustd[ilat, ilon] = np.nanstd(drifters['u'][box])
Vstd[ilat, ilon] = np.nanstd(drifters['v'][box])
[5]:
cenlon
[5]:
array([[165.5, 166. , 166.5, ..., 266.5, 267. , 267.5],
[165.5, 166. , 166.5, ..., 266.5, 267. , 267.5],
[165.5, 166. , 166.5, ..., 266.5, 267. , 267.5],
...,
[165.5, 166. , 166.5, ..., 266.5, 267. , 267.5],
[165.5, 166. , 166.5, ..., 266.5, 267. , 267.5],
[165.5, 166. , 166.5, ..., 266.5, 267. , 267.5]])
[6]:
cenlat
[6]:
array([[-9.5, -9.5, -9.5, ..., -9.5, -9.5, -9.5],
[-9. , -9. , -9. , ..., -9. , -9. , -9. ],
[-8.5, -8.5, -8.5, ..., -8.5, -8.5, -8.5],
...,
[ 8.5, 8.5, 8.5, ..., 8.5, 8.5, 8.5],
[ 9. , 9. , 9. , ..., 9. , 9. , 9. ],
[ 9.5, 9.5, 9.5, ..., 9.5, 9.5, 9.5]])
And now let’s plot then
[7]:
fig, ax = plt.subplots(2,1,figsize=(15,8), sharex=True)
q = ax[0].quiver(cenlon, cenlat, Umean, Vmean)
ax[0].quiverkey(q, 178, 11, 0.25, '0.25 m/s', coordinates='data')
ax[0].set_ylabel('Latitude')
ax[0].set_xlabel('Longitude')
ax[0].set_title('Mean currents Equatorial Pacific from WOCE drifters')
ax[0].set_aspect('equal')
ax[0].set_ylim(-10, 12)
cs = ax[1].contourf(cenlon, cenlat, np.sqrt(Ustd**2 + Vstd**2), levels=50)
ax[1].set_aspect('equal')
cbaxes = fig.add_axes([0.11, 0.05, 0.8, 0.03])
cb = plt.colorbar(cs, cax = cbaxes, orientation='horizontal')

Now let’s try making zonal strips
[8]:
cenlat1 = np.arange(-9.5, 9.6, 0.5)
Umean1 = np.nan * np.ones_like(cenlat1)
Vmean1 = np.nan * np.ones_like(cenlat1)
Ustd1 = np.nan * np.ones_like(cenlat1)
Vstd1 = np.nan * np.ones_like(cenlat1)
for ilat in range(len(cenlat1)):
strip = np.where(np.absolute(drifters['lat']-cenlat1[ilat]) < 0.5)[0]
Umean1[ilat] = np.nanmedian(drifters['u'][strip])
Vmean1[ilat] = np.nanmedian(drifters['v'][strip])
Ustd1[ilat] = np.nanstd(drifters['u'][strip])
Vstd1[ilat] = np.nanstd(drifters['v'][strip])
And plot them
[10]:
fig, ax = plt.subplots(1,4,figsize=(15,12), sharey=True)
q = ax[0].quiver(np.zeros_like(cenlat1), cenlat1, Umean1, Vmean1)
ax[0].quiverkey(q, -178, 11, 0.25, '0.25 m/s', coordinates='data')
ax[0].set_ylabel('Latitude')
ax[0].set_title('Mean currents from drifters')
#ax[0].set_aspect('equal')
ax[0].set_ylim(-10, 12)
cs = ax[1].plot(Vmean1, cenlat1, 'k')
ax[1].set_title('meridional current')
cs = ax[2].plot(Umean1, cenlat1, 'k')
ax[2].set_title('zonal current')
cs = ax[3].plot(np.sqrt(Ustd1**2+Vstd1**2), cenlat1, 'k')
ax[3].set_title('standard deviation')
[10]:
Text(0.5, 1.0, 'standard deviation')

Pacific Variability- ENSO¶
There are several different indices for ENSO: https://climatedataguide.ucar.edu/climate-data/nino-sst-indices-nino-12-3-34-4-oni-and-tni.
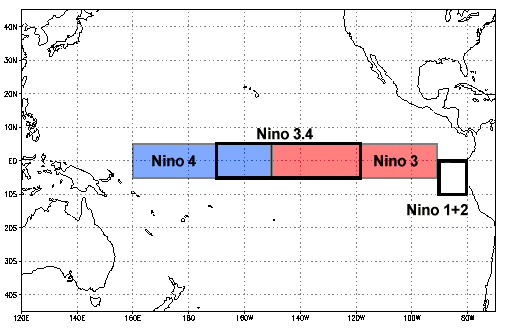
We use the Nino 3.4 index. The data are easily found online:
[11]:
ninocols = ['year', 'month', 'sst', 'ssta', 'anom']
nino = pd.read_csv('https://www.cpc.ncep.noaa.gov/products/analysis_monitoring/ensostuff/detrend.nino34.ascii.txt',
names=ninocols, delim_whitespace=True, skiprows=1)
[12]:
nino
[12]:
| year | month | sst | ssta | anom | |
|---|---|---|---|---|---|
| 0 | 1950 | 1 | 24.56 | 26.18 | -1.62 |
| 1 | 1950 | 2 | 25.07 | 26.39 | -1.32 |
| 2 | 1950 | 3 | 25.88 | 26.95 | -1.07 |
| 3 | 1950 | 4 | 26.29 | 27.39 | -1.11 |
| 4 | 1950 | 5 | 26.19 | 27.56 | -1.37 |
| ... | ... | ... | ... | ... | ... |
| 861 | 2021 | 10 | 25.77 | 26.72 | -0.94 |
| 862 | 2021 | 11 | 25.76 | 26.70 | -0.94 |
| 863 | 2021 | 12 | 25.54 | 26.60 | -1.07 |
| 864 | 2022 | 1 | 25.60 | 26.55 | -0.95 |
| 865 | 2022 | 2 | 25.87 | 26.76 | -0.89 |
866 rows × 5 columns
We need to fix the date; simplest way forward is:
[13]:
date = nino['year'] + nino['month']/12
Let’s plot all the temperatures and anomalies, using the sharex argumnet to clean up the x-axes.
[14]:
fig, ax = plt.subplots(3,1,figsize=(18,12), sharex=True)
ax[0].plot(date, nino['sst'], 'k')
ax[0].set_title('Nino 3.4 temperatures')
ax[1].plot(date, nino['ssta'], 'k')
ax[1].set_title('Nino 3.4 temperatures, climate-adjusted')
ax[2].plot(date, nino['anom'], 'k')
ax[2].set_title('Nino 3.4 anomalies')
[14]:
Text(0.5, 1.0, 'Nino 3.4 anomalies')

Let’s zoom on last 30 years
[15]:
fig, ax = plt.subplots(3,1,figsize=(18,12), sharex=True)
ax[0].plot(date, nino['sst'], 'k')
ax[0].set_title('Nino 3.4 temperatures')
ax[1].plot(date, nino['ssta'], 'k')
ax[1].set_title('Nino 3.4 climate-adjusted temperatures')
ax[2].plot(date, nino['anom'], 'k')
ax[2].set_title('Nino 3.4 anomalies')
for axis in ax:
axis.set_xlim(1990, 2021.25)

Below is the code used to make the drifter file; you do not need to run it, but if you want to get a different sub-set of the data, you can change it to a code cell, modify it, and use it.
import pandas as pd import numpy as np import datetime # dates in days since 1 Jan year 1AD mindate, maxdate = datetime.datetime(1900,1,1).toordinal(), datetime.datetime(2022,1,1).toordinal() minlat, maxlat = -10, 10 minlon, maxlon = 165, 360-92 urls = ['ftp://ftp.aoml.noaa.gov/pub/phod/buoydata/buoydata_1_5000.dat.gz', 'ftp://ftp.aoml.noaa.gov/pub/phod/buoydata/buoydata_5001_10000.dat.gz', 'ftp://ftp.aoml.noaa.gov/pub/phod/buoydata/buoydata_10001_15000.dat.gz'] # list of names of columns colnames = ['id','month','day','year','lat','lon','sst','u','v','umag','latvar','lonvar','tempvar'] # make empty dataframe for appending just the data we want to keep to gooddata = pd.DataFrame([]) for ifile in urls: data = pd.read_csv(ifile, compression='gzip', delim_whitespace=True, names=colnames) # make date into ordinal number, days since 1 Jan year 1 date = np.array([datetime.datetime(yy, mm, int(dd)).toordinal() + dd%1 for yy, mm, dd in zip(data['year'], data['month'], data['day'])]) # find indices with each limit imindate = np.where(date>=mindate)[0] imaxdate = np.where(date<=maxdate)[0] iminlat = np.where(data['lat']>=minlat)[0] imaxlat = np.where(data['lat']<=maxlat)[0] iminlon = np.where(data['lon']>=minlon)[0] imaxlon = np.where(data['lon']<=maxlon)[0] # combine the indices, keeping only ones that satisfy both limits idate = set(imindate).intersection(imaxdate) ilat = set(iminlat).intersection(imaxlat) ilon = set(iminlon).intersection(imaxlon) # combine date, lat, and lon limits into a single list of indices # delete extra variables idata = sorted(list((idate.intersection(ilat)).intersection(ilon))) del imindate, imaxdate, iminlat, imaxlat, iminlon, imaxlon del date, idate, ilat, ilon # append data to new dataframe gooddata = gooddata.append(data.iloc[idata]) # save our new dataframe as its own csv file with gzip compression #gooddata.to_csv('/home/ocn463-data/woce/drift/pacdrift.gz', compression='gzip', index=False)[ ]: